

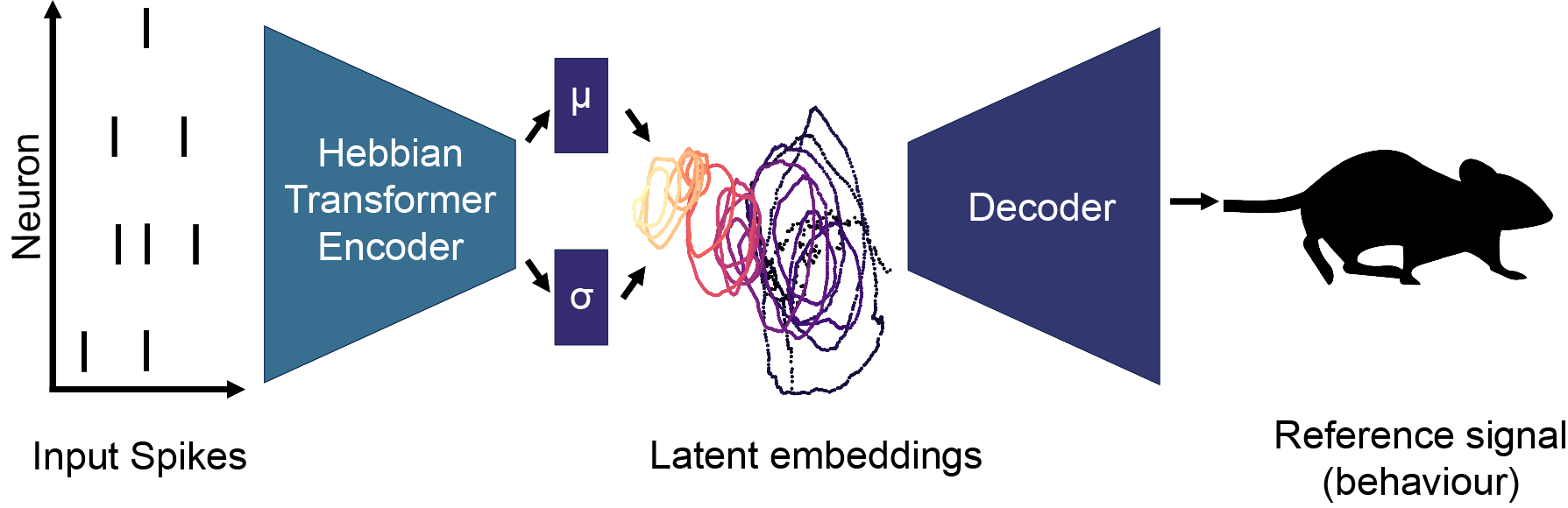
Two-dimensional embeddings obtained during the supervised prediction of the position of the hand of monkeys performing a centre-out task (Chowdury et al., 2020).
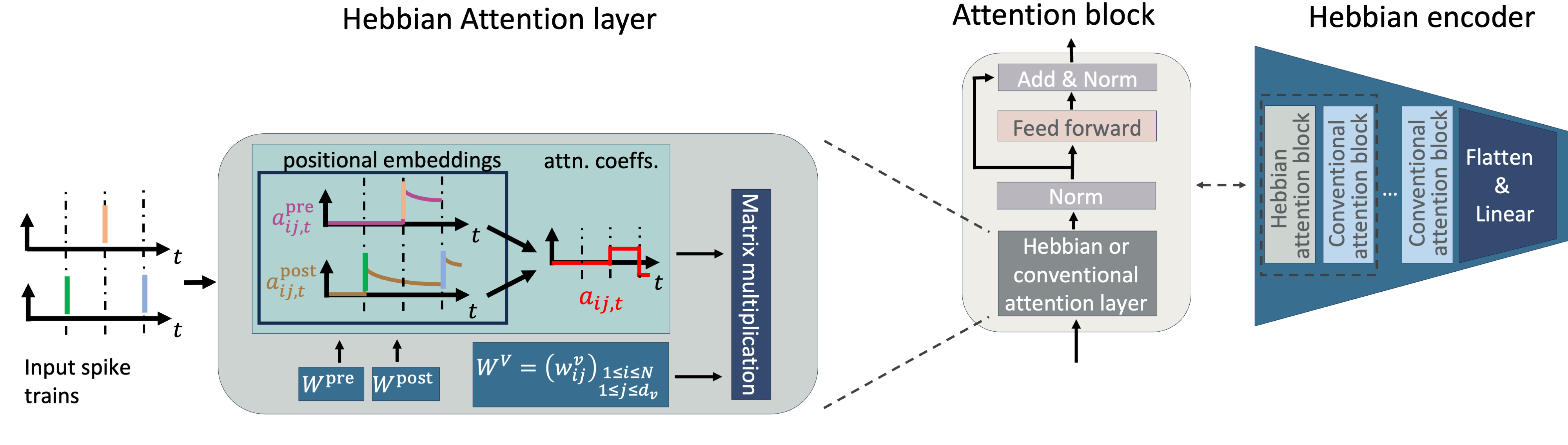
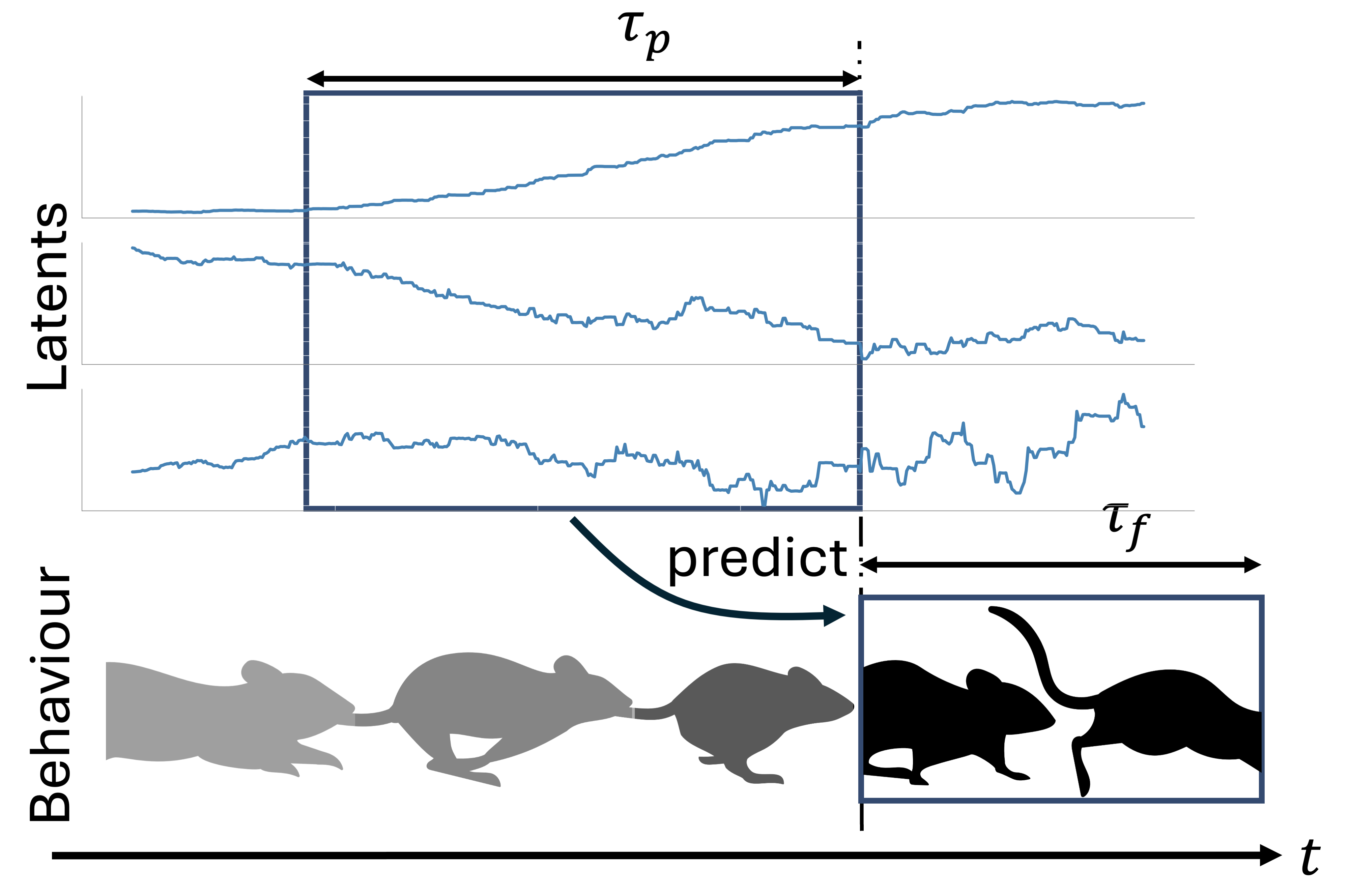
Understanding how the brain represents sensory information and triggers behavioural responses is a fundamental goal in neuroscience. Recent advances in neuronal recording techniques aim to progress towards this milestone, yet the resulting high-dimensional responses are challenging to interpret and link to relevant variables. Although existing machine learning models propose to do so, they often sacrifice interpretability for predictive power, effectively operating as black boxes. In this work, we introduce SPARKS, a biologically inspired model capable of high decoding accuracy and interpretable discovery within a single framework. SPARKS adapts the self-attention mechanism of large language models to extract information from the timing of single spikes and the sequence in which neurons fire using Hebbian learning. Trained with a criterion inspired by predictive coding to enforce temporal coherence, our model produces low-dimensional latent embeddings that are robust across sessions and animals. By directly capturing the underlying data distribution through a generative encoding-decoding framework, SPARKS exhibits state-of-the-art predictive capabilities across diverse electrophysiology and calcium imaging datasets from the motor, visual and entorhinal cortices. Crucially, the Hebbian coefficients learned by the model are interpretable, allowing us to infer the effective connectivity and recover the known functional hierarchy of the mouse visual cortex. Overall, SPARKS unifies representation learning, high-performance decoding and model interpretability in a single framework by bridging neuroscience and AI, providing a powerful and versatile tool for dissecting neural computations and marking a step towards the next generation of biologically inspired intelligent systems.
@article {skatchkovsky24sparks,
author = {Skatchkovsky, Nicolas and Glazman, Natalia and Sadeh, Sadra and Iacaruso, Florencia},
title = {A Biologically Inspired Attention Model for Neural Signal Analysis},
elocation-id = {2024.08.13.607787},
year = {2024},
doi = {10.1101/2024.08.13.607787},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/08/16/2024.08.13.607787},
eprint = {https://www.biorxiv.org/content/early/2024/08/16/2024.08.13.607787.full.pdf},
journal = {bioRxiv}
}